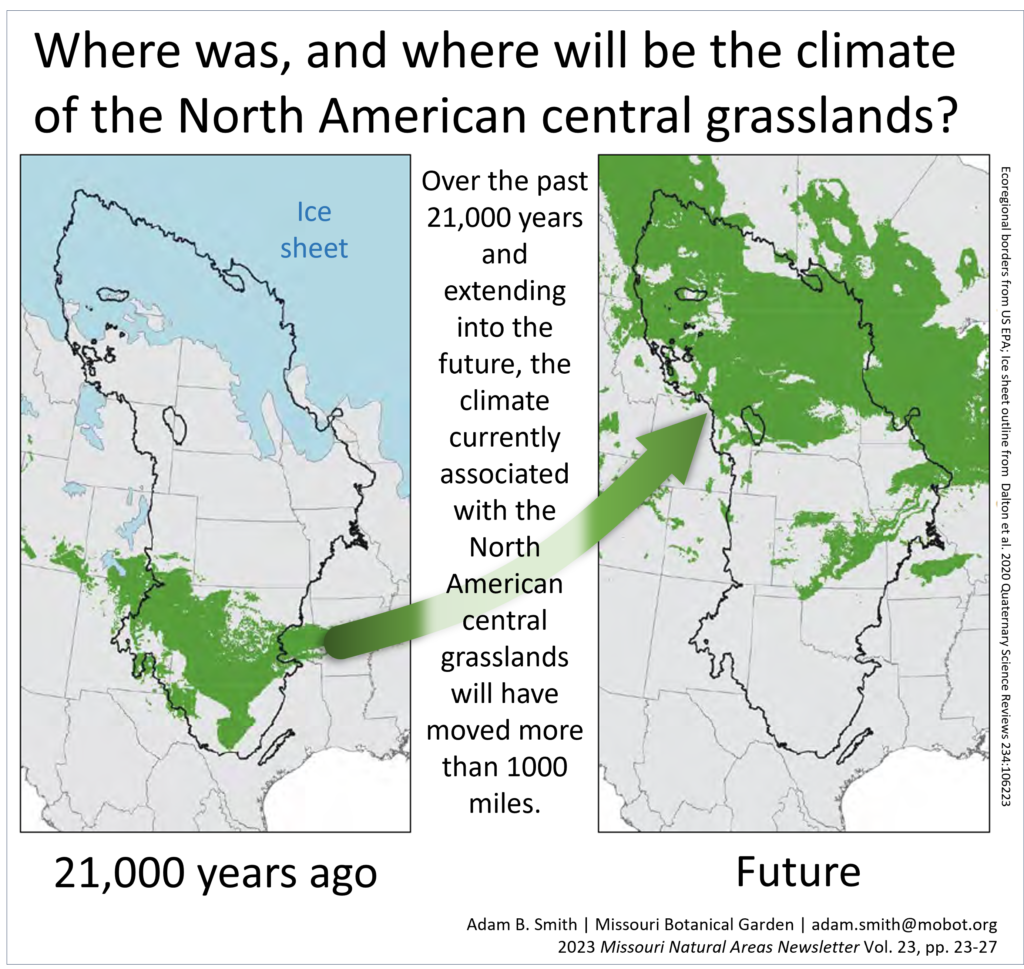
Recently Adam was asked to pen a thought-piece for the Missouri Natural Area Newsletter about climate change in the Midwest. In 5-1/2 easy pages, it covers: emissions scenarios, the last glacial maximum, the year 2500, and how we need an adaptive resist-adapt-direct (RAD) framework for dealing with it. Where will Midwest ecosystems be in 2500? …
Continue reading Where will Midwestern ecosystems be in the year 2500?
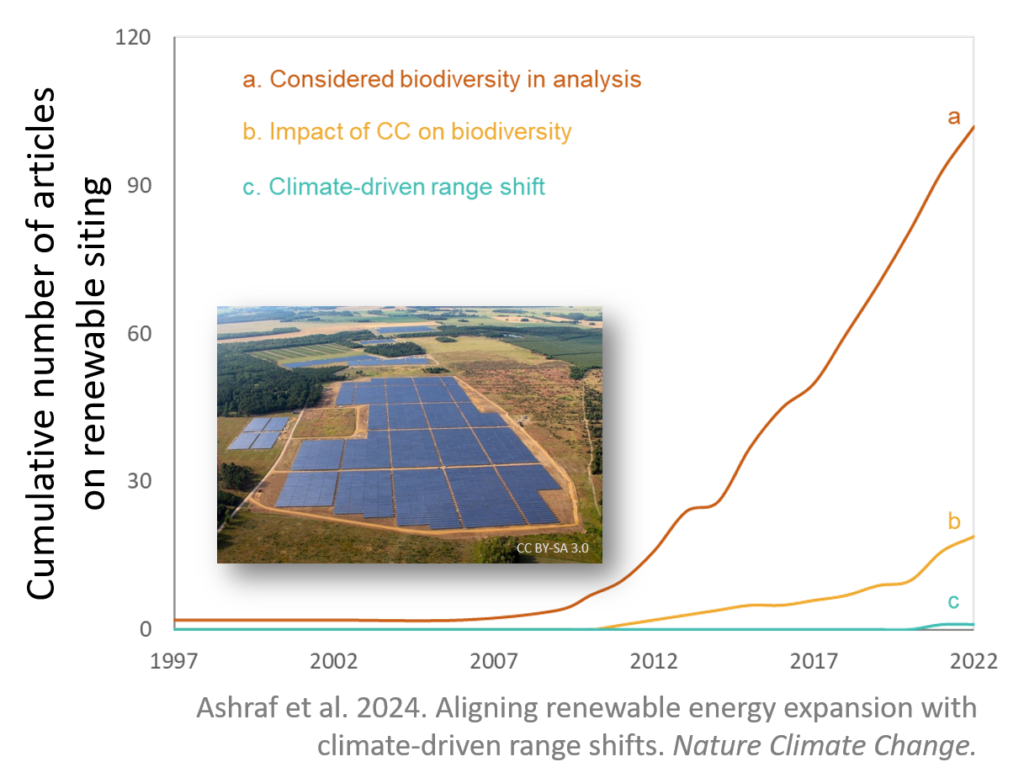
Renewable energy versus(?) biodiversity
Renewable energy might save the world, but we need to be sure it doesn’t destroy it along the way. . . Unless located and designed wisely, solar and wind installations could actually harm species as they shift in response to climate change. Owing to their large size, large-scale renewable energy plants have an outsized impact …
Continue reading Renewable energy versus(?) biodiversity
Join us in a study of SDM methodology!
We invite you to be part of a worldwide study about structural uncertainty in species distribution models (SDMs)! Specifically, we are interested in how choices along the modeling workflow affect final outcomes of models. While differences in algorithms, spatial extents, etc., have been widely explored, we are interested in the cumulative effect of choices and …
Continue reading Join us in a study of SDM methodology!
Thank you, Postcard Underground!
Having chosen the path of a scientist who studies climate change so that I could do things with my life to make this world a better place, I never expected to be criticized for being a liar, greedy person, or dupe. Support like this helps. Thank you, Postcard Underground. You know who you are 😉
Integrating occurrences, pollen, and DNA to reconstruct species’ biogeographic histories
To date, most reconstructions of species’ biogeographic histories have relied on a single data source, such as occurrence data analyzed using species distribution models, fossil pollen analyzed using pollen-vegetation models, or genetics analyzed using scenario-based modeling. “Integration” between them has largely been “by eye.” We present a statistically integrated model based on approximate Bayesian computation …
Continue reading Integrating occurrences, pollen, and DNA to reconstruct species’ biogeographic histories
Paid summer research on the effect of climate change on plant reproduction and physiology at the Missouri Botanical Garden
Drs. Matt Austin and Adam Smith are excited to recruit a participant for the summer 2024 session of Missouri Botanical Garden’s Research Experience for Undergraduates (REU) Program sponsored by the National Science Foundation. Contemporary climate change has already altered many aspects of the natural world, including the timing of reproduction and physiological requirements. Many plants …
Continue reading Paid summer research on the effect of climate change on plant reproduction and physiology at the Missouri Botanical Garden
We’re back!
Website looks a little empty, doesn’t it? It crashed. :/ But we do good science with good people for a good planet.